当你的肿瘤高通量测序样本越多,或没有病人自身配对的癌旁与血液等非体细胞突变受累组织的配对时,那么WES或WGS所获得的体细胞突变数量将会非常多,所涉及的基因也很多,以至于突变Maf图无法阅读 (基因太多、图太大)。类似这样:
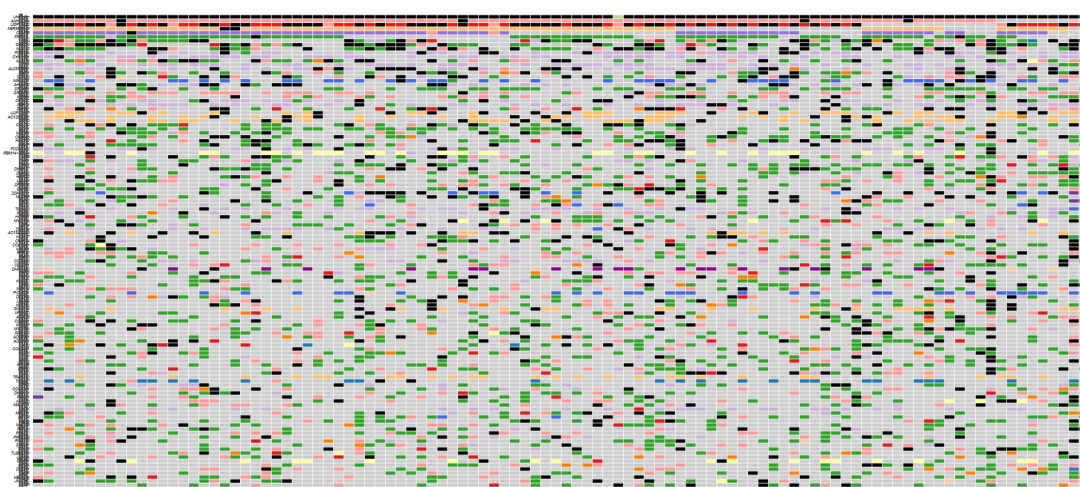
R包 maftools绘制的100+肿瘤样本突变图景
你所关注的肿瘤驱动突变就在这个大图里。但怎么过滤、筛选,是肿瘤测序分析的另一个难点,甚至是核心内容了。第1个难点在于GATK变异检测后的质控与过滤。变异注释、Maf格式整理也有需要注意的地方。 进一步筛选的原则是:做一些初步的、逐步的、不丢失核心信息的筛选,使图不至于过大 (可以很大,但不能过大;也不能一下就变得很小,否则会丢失重要的、可能的肿瘤驱动基因)。 这套筛选的逻辑与原则,跟转录组中首先依赖 Fold Change 和 P值 等指标筛选差异基因不同。肿瘤基因组分析似乎是另一整套筛选逻辑,核心是看变异的功能效应,所谓的“肿瘤驱动”也属于这个范围。
当然,肿瘤高通量测序的QC (质量控制)部分的筛选也很重要。其任务是:相对准确地挑出真实的体细胞突变,而不理会突变的功能影响。由于肿瘤自身突变的异质性特点,以及取样纯度的客观因素限制,驱动突变的负荷很低 (0.5%~10%),甚至与测序错误相接近,因此这里的筛选 (挑出真实的体细胞突变,完美过滤掉遗传变异)在统计学上是极难的,甚至是不可能完美解决的。 但如本节开头所述,即使都是体细胞突变,所涉及的基因个数也是非常多、且杂乱的,需要进一步结合“突变功能效应” (比如:同义、错义突变,剪接位点,内含子、基因间,药物响应、耐药性等),做更加深入、核心的筛选。假设你只关注EGFR的各种突变,那问题就非常简单了,可在当前最大的那个Maf图里 (即使图太大、无法阅读),只需取出EGFR基因即可。
肿瘤突变功能筛选的几个可选技术路径
① 疾病名称或关键词
这个思路是为了看:在已经报道的该癌种的众多突变 (要尽量收集全),是否在当前的肿瘤研究项目中出现。
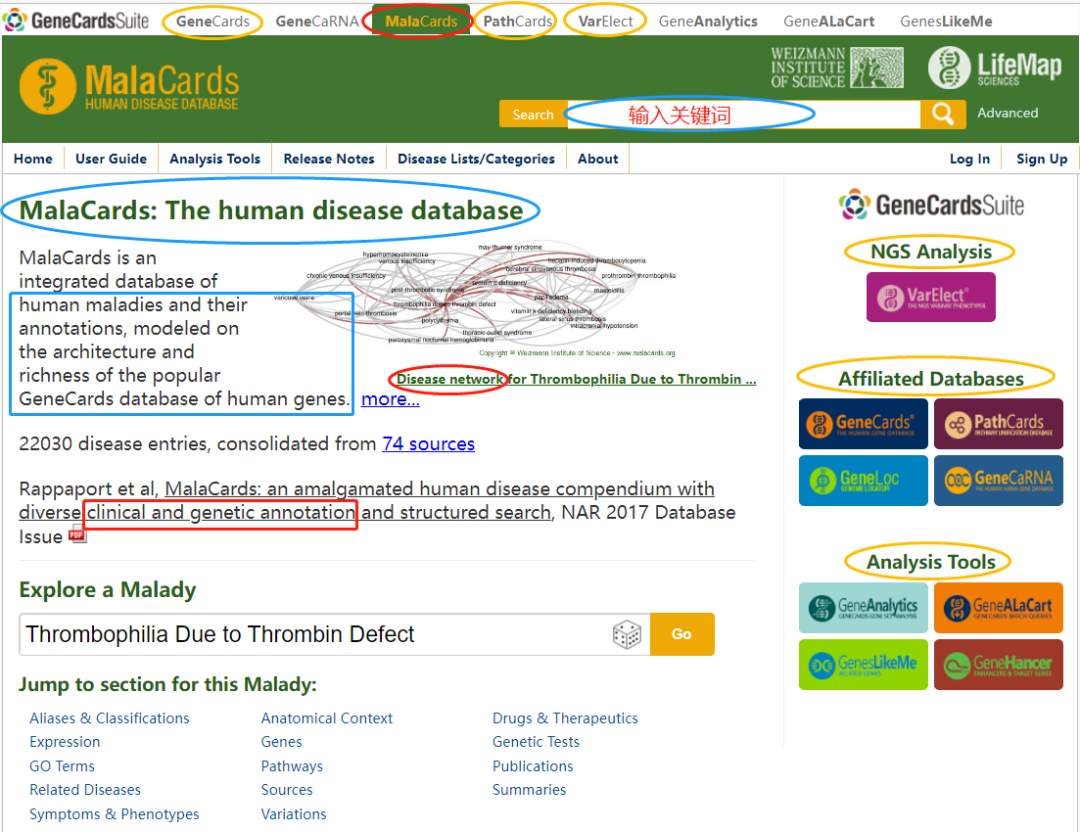
如脑胶质瘤的疾病关键词:Glioma,Glioblastoma,Low grade glioma,High grade glioma,Diffuse Astrocytoma (看起来Glioma可能囊括其它的,故可先选用之)。为什么我知道这么多疾病的别名?搜一下MalaCards。
② 汇总该疾病已知的基因检测Panel
http://www.zg-bio.com/content/index/classid/11/id/22 (真固生物)
是一种细胞增殖相关的核抗原,作为判定增殖细胞数比例的指标,Ki-67蛋白存在于其染色阳性说明癌细胞增殖活跃,阳性标记指数越高,则恶性程度越高,预后越差,可通过免疫组化来检测。 异柠檬酸脱氢酶(IDH)突变在原发性胶质母细胞瘤中发生率很低,有突变的患者治疗效果和预后更好。可以用来鉴别胶质瘤和胶质细胞增生,有无IDH1/2基因突变作为评估低级别胶质瘤患者风险级别的指标之一。 MGMT基因启动子CpG岛甲基化在判断脑胶质瘤患者预后及预测肿瘤对烷化剂药物耐药性方面具有重要意义。具有MGMT启动子甲基化的胶质瘤患者对放疗和化疗更敏感,并具有更长的生存期。 染色体1p/19q联合性缺失是指1号染色体短臂和19号染色体长臂同时缺失,目前认为1p/19q联合性缺失是少突胶质细胞瘤的分子特征,是其诊断性分子标志物。对于有1p/19q联合缺失的少突或间变少突胶质细胞瘤患者,推荐化疗或联合放化疗。1p/19q联合缺失的胶质瘤患者总生存期和无进展生存期较长。 端粒酶逆转录酶(TelomeraseReverseTranscriptase,TERT)是端粒酶复合物的催化中心,最新研究表明:只携带TERT突变的III-IV级胶质瘤患者多为原发性胶母细胞瘤,且预后不良;只携带IDH1/2突变的III-IV级胶质瘤患者多呈现星形细胞形态;同时携带TERT和IDH1/2突变的胶质瘤患者多呈现少突胶质细胞形态,预后良好。 BRAF基因位于7q34,编码一种丝/苏氨酸特异性激酶,近年临床试验表明,维莫非尼(vemurafenib)在儿童胶质母细胞瘤、毛细胞黏液样星形细胞瘤、复发多形性星形细胞瘤等的治疗中均也取得了较好的疗效,提示BRAF突变的患者可选取维莫非尼可作为潜在靶向的治药物。 TP53为抑癌基因,定位于染色体17p13.1,编码蛋白称为p53蛋白,p53蛋白能调节细胞周期和避免细胞癌变发生,超过50%的人类肿瘤涉及TP53基因突变的发生。TP53基因突变在低级别星形细胞瘤中发生率为50%-60%,继发性GBM发生率为70%,原发性GBM发生率为25%-37%。目前p53蛋白可通过免疫组化检测。基因水平可通过PCR测序检测TP53突变。建议:TP53突变在低级别星形细胞瘤和继发性GBM中发生率高,有TP53突变的低级别胶质瘤预后较差。 组蛋白(histone)常有多种变体,共分为5种亚基,分别为H1、H2A、H2B、H3和H4。主要参与基因表达的精细化调节,具有调节方式多种、不同变体具有不同的作用。2016年WHO中枢神经系统肿瘤分类中将其单独分为一个新的类型。组蛋白H3.3突变中线结构(如丘脑、脑干及脊髓等)区域胶质瘤中具有极高的表达,且常见于儿童和年轻成人,呈弥漫性生长,肿瘤恶性程度极高,预后极差。相关研究均显示H3K27M突变在弥漫性中线胶质瘤中独特的基因突变模式。潜在获益药物ONC201,Valproicacid. 磷酸酯酶与张力蛋白同源物(PTEN)定位于染色体10q23.3,是蛋白质络氨酸磷酸酶基因家族成员,是重要的抑癌基因。PTEN蛋白是磷酸酶,它使蛋白质去磷酸化而发挥作用,参与信号通路的转导,在细胞生长/分裂的速度过快或分裂不受控制时,能够调控细胞分裂周期,使细胞停止分裂并诱导凋亡,这些功能可阻止细胞的异常增殖进而限制肿瘤的形成。建议对WHOIII级和IV级的胶质瘤样本检测PTEN的突变。有PTEN突变的间变星形细胞瘤患者预后较差。https://zhuanlan.zhihu.com/p/163208947
EGFR扩增和EGFRv III重排
实验室检测方法:
EGFR扩增:荧光原位杂交;
EGFRvⅢ重排:实时定量PCR,免疫组织化学,多重探针依赖式扩增技术。推荐使用荧光原位杂交检测EGFR重排。
建议:有EGFR扩增的大于60岁的GBM患者预后差,诊断方面的意义表现在两方面:对小细胞GBM的诊断;辅助判定活检组织的病理结果。
miR-181d
实验室检测方法:原位杂交。
建议:miR-181d对于GBM是一个预测预后的可靠指标。临床检测miR-181d的表达水平能提示GBM患者对TMZ化疗的敏感性。MIR181D
https://zhuanlan.zhihu.com/p/28493988
cut -f 1 all.maf | grep -i miRMIR1-1HG-AS1MIR548YMIR3663HGMIR4495MIR3663HGMIR8088MIR4267MIR4444-2MIR3689D2MIR1-1HG
③ 以生物通路、生物过程,甚至自定义的目标基因的集合来筛选
④ 按照突变的“功能效应”筛选
例如:同义/错义突变,剪接位点影响,处在内含子或基因间,对已获批药物的明确的响应与耐药性,等等。
⑤ 依赖数据库文件
将GATK Funcotator注释的all.maf文件 (217列注释信息,非常全,如下),与COSMIC (Catalogue of Somatic Mutations in Cancer,癌症体细胞突变目录)、ClinVar变异注释库文件 (variant_summary_GRCh38.bed.txt)合并,再以特定癌种发病机制中的关键词检索,获得目标基因及其变异位点。GATK Funcotator注释的all.maf文件(列的名称)
Hugo_Symbol Entrez_Gene_Id Center NCBI_Build Chromosome Start_Position End_Position Strand Variant_Classification Variant_Type Reference_Allele Tumor_Seq_Allele1 Tumor_Seq_Allele2 dbSNP_RS dbSNP_Val_Status Tumor_Sample_Barcode Matched_Norm_Sample_Barcode Match_Norm_Seq_Allele1 Match_Norm_Seq_Allele2 Tumor_Validation_Allele1 Tumor_Validation_Allele2 Match_Norm_Validation_Allele1 Match_Norm_Validation_Allele2 Verification_Status Validation_Status Mutation_Status Sequencing_Phase Sequence_Source Validation_Method Score BAM_File Sequencer Tumor_Sample_UUID Matched_Norm_Sample_UUID Genome_Change Annotation_Transcript Transcript_Strand Transcript_Exon Transcript_Position cDNA_Change Codon_Change Protein_Change Other_Transcripts Refseq_mRNA_Id Refseq_prot_Id SwissProt_acc_Id SwissProt_entry_Id Description UniProt_AApos UniProt_Region UniProt_Site UniProt_Natural_Variations UniProt_Experimental_Info GO_Biological_Process GO_Cellular_Component GO_Molecular_Function COSMIC_overlapping_mutations COSMIC_fusion_genes COSMIC_tissue_types_affected COSMIC_total_alterations_in_gene Tumorscape_Amplification_Peaks Tumorscape_Deletion_Peaks TCGAscape_Amplification_Peaks TCGAscape_Deletion_Peaks DrugBank ref_context gc_content CCLE_ONCOMAP_overlapping_mutations CCLE_ONCOMAP_total_mutations_in_gene CGC_Mutation_Type CGC_Translocation_Partner CGC_Tumor_Types_Somatic CGC_Tumor_Types_Germline CGC_Other_Diseases DNARepairGenes_Activity_linked_to_OMIM FamilialCancerDatabase_Syndromes MUTSIG_Published_Results OREGANNO_ID OREGANNO_Values tumor_f t_alt_count t_ref_count n_alt_count n_ref_count Gencode_34_secondaryVariantClassification Achilles_Top_Genes ClinVar_VCF_AF_ESP ClinVar_VCF_AF_EXAC ClinVar_VCF_AF_TGP ClinVar_VCF_ALLELEID ClinVar_VCF_CLNDISDB ClinVar_VCF_CLNDISDBINCL ClinVar_VCF_CLNDN ClinVar_VCF_CLNDNINCL ClinVar_VCF_CLNHGVS ClinVar_VCF_CLNREVSTAT ClinVar_VCF_CLNSIG ClinVar_VCF_CLNSIGCONF ClinVar_VCF_CLNSIGINCL ClinVar_VCF_CLNVC ClinVar_VCF_CLNVCSO ClinVar_VCF_CLNVI ClinVar_VCF_DBVARID ClinVar_VCF_GENEINFO ClinVar_VCF_MC ClinVar_VCF_ORIGIN ClinVar_VCF_RS ClinVar_VCF_SSR ClinVar_VCF_ID ClinVar_VCF_FILTER CosmicFusion_fusion_id Familial_Cancer_Genes_Synonym Familial_Cancer_Genes_Reference Gencode_XHGNC_hgnc_id HGNC_HGNC_ID HGNC_Status HGNC_Locus_Type HGNC_Locus_Group HGNC_Previous_Symbols HGNC_Previous_Name HGNC_Synonyms HGNC_Name_Synonyms HGNC_Chromosome HGNC_Date_Modified HGNC_Date_Symbol_Changed HGNC_Date_Name_Changed HGNC_Accession_Numbers HGNC_Enzyme_IDs HGNC_Ensembl_Gene_ID HGNC_Pubmed_IDs HGNC_RefSeq_IDs HGNC_Gene_Family_ID HGNC_Gene_Family_Name HGNC_CCDS_IDs HGNC_Vega_ID HGNC_OMIM_ID(supplied_by_OMIM) HGNC_RefSeq(supplied_by_NCBI) HGNC_UniProt_ID(supplied_by_UniProt) HGNC_Ensembl_ID(supplied_by_Ensembl) HGNC_UCSC_ID(supplied_by_UCSC) Oreganno_Build Simple_Uniprot_alt_uniprot_accessions dbSNP_ASP dbSNP_ASS dbSNP_CAF dbSNP_CDA dbSNP_CFL dbSNP_COMMON dbSNP_DSS dbSNP_G5 dbSNP_G5A dbSNP_GENEINFO dbSNP_GNO dbSNP_HD dbSNP_INT dbSNP_KGPhase1 dbSNP_KGPhase3 dbSNP_LSD dbSNP_MTP dbSNP_MUT dbSNP_NOC dbSNP_NOV dbSNP_NSF dbSNP_NSM dbSNP_NSN dbSNP_OM dbSNP_OTH dbSNP_PM dbSNP_PMC dbSNP_R3 dbSNP_R5 dbSNP_REF dbSNP_RV dbSNP_S3D dbSNP_SAO dbSNP_SLO dbSNP_SSR dbSNP_SYN dbSNP_TOPMED dbSNP_TPA dbSNP_U3 dbSNP_U5 dbSNP_VC dbSNP_VP dbSNP_WGT dbSNP_WTD dbSNP_dbSNPBuildID dbSNP_ID dbSNP_FILTER HGNC_Entrez_Gene_ID(supplied_by_NCBI) dbSNP_RSPOS dbSNP_VLD AS_FilterStatus AS_SB_TABLE AS_UNIQ_ALT_READ_COUNT CONTQ DP ECNT GERMQ MBQ MFRL MMQ MPOS NALOD NCount NLOD OCM PON POPAF ROQ RPA RU SEQQ STR STRANDQ STRQ TLODClinVar变异注释库文件(列的名称)
Chromosome Start Stop #AlleleID Type Name GeneID GeneSymbol HGNC_ID ClinicalSignificance ClinSigSimple LastEvaluated RS# (dbSNP) nsv/esv (dbVar) RCVaccession PhenotypeIDS PhenotypeList Origin OriginSimple Assembly ChromosomeAccession Chromosome Start Stop ReferenceAllele AlternateAllele Cytogenetic ReviewStatus NumberSubmitters Guidelines TestedInGTR OtherIDs SubmitterCategories VariationID PositionVCF ReferenceAlleleVCF AlternateAlleleVCF 可能有其它更多筛选思路。这里列举的5个思路都可以逐步尝试一下,看哪个效果合适。目的是:突变瀑布图可被阅读,契合研究的目标,等等。当然,这几个思路也可以组合起来使用。 以脑胶质瘤为例,选择“Glioma”为关键词 (原因上文已经描述),使用的工具是MalaCards:第6期 | 临床基因组/外显组数据分析实战 (课件)

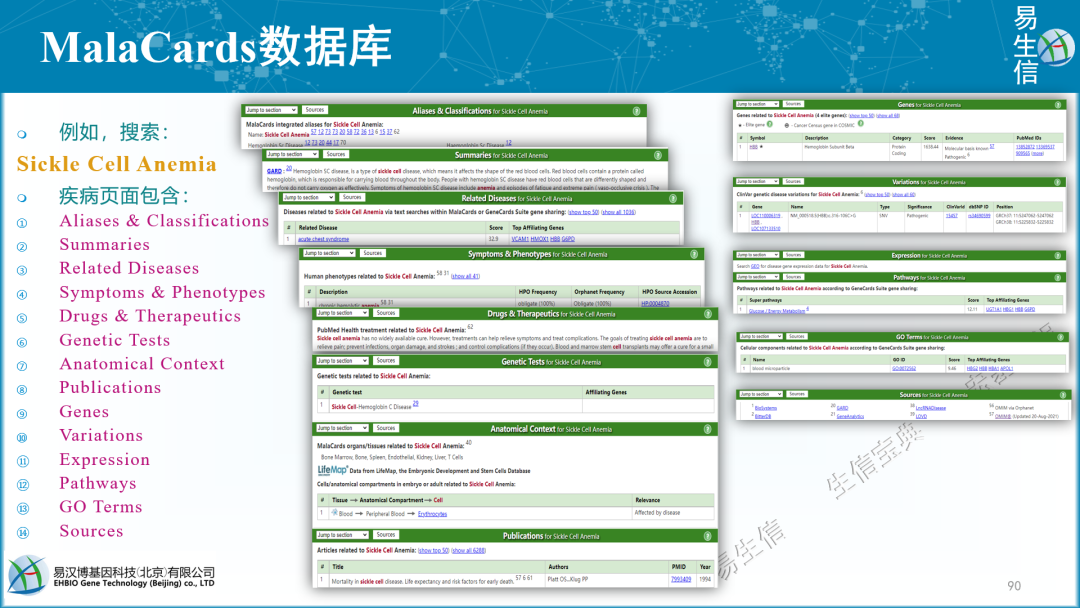
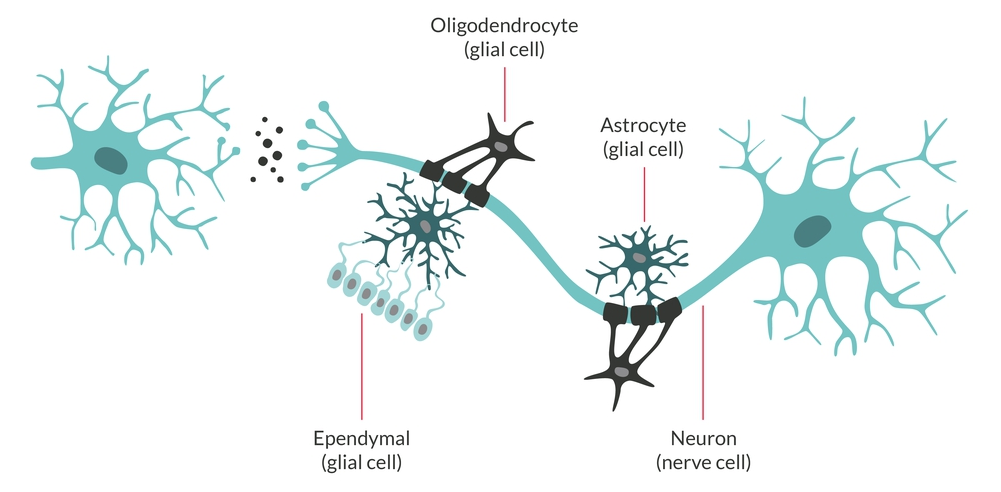
https://www.malacards.org/GARD (红体字是将被选取、用于生信筛选的关键词) 胶质瘤 (Glioma)是指从神经胶质细胞 (Glial cell)发展而来的一种脑部肿瘤。神经胶质细胞本身是围绕、支持大脑中神经元 (即神经细胞。Neuron: Nerve cell)的特化的细胞。
胶质瘤通常根据肿瘤中所涉及的神经胶质细胞类型进行分类:① 星形细胞瘤 (Astocytoma) - 由称为星形胶质细胞的星形神经胶质细胞发展而来的肿瘤;② 室管膜瘤 (Ependymoma) - 由排列在脑室和脊髓中心的室管膜细胞产生的肿瘤;③ 少突胶质细胞瘤 (Oligodendroglioma) - 影响少突胶质细胞的肿瘤。
胶质瘤的症状因类型而异,但可能包括:头痛,恶心和呕吐,混乱,性格变化,平衡问题,视力问题,言语困难 (和/或癫痫发作)。胶质瘤确切的根本原因尚不清楚。在大多数情况下,肿瘤在没有家族病史的人身上偶尔发生 (即起源于随机的体细胞突变)。治疗取决于许多因素,包括:肿瘤的类型、大小、阶段和位置;可能包括:手术、放射疗法、化学疗法 (和/或靶向疗法)。
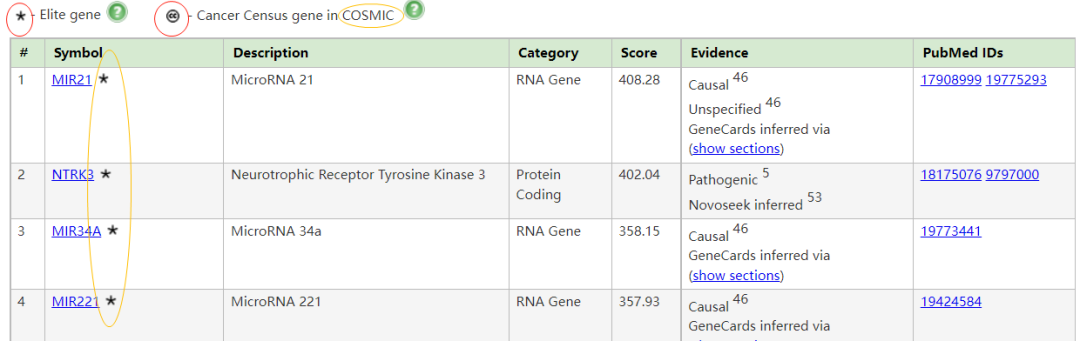
胶质瘤与高级别胶质瘤 (High grade glioma)和胶质母细胞瘤 (Glioblastoma)有关。与胶质瘤相关的一个重要基因是MIR21 (MicroRNA 21),其相关途径/超级通路 (Superpathway)包括:细胞分化扩胀指数 (Cell differentiation - expanded index);参与DNA损伤反应的miRNA (miRNAs involved in DNA damage response)。在这种疾病的背景下,已经提到了药物达布拉非尼和乳糖醇 (Dabrafenib and Lactitol)。有关连的组织包括:大脑、脊髓和T细胞。
Inherited polymorphisms of the DNA repair genes
Germ-line (inherited) polymorphisms of the DNA repair genes ERCC1, ERCC2 (XPD) and XRCC1 increase the risk of glioma. This indicates that altered or deficient repair of DNA damage contributes to the formation of gliomas. DNA damages are a likely major primary cause of progression to cancer in general.
Excess DNA damages can give rise to mutations through translesion synthesis. Furthermore, incomplete DNA repair can give rise to epigenetic alterations or epimutations. Such mutations and epimutations may provide a cell with a proliferative advantage which can then, by a process of natural selection, lead to progression to cancer.
Epigenetic repression of DNA repair genes is often found in progression to sporadic glioblastoma. For instance, methylation of the DNA repair gene MGMT promoter was observed in 51% to 66% of glioblastoma specimens. In addition, in some glioblastomas, the MGMT protein is deficient due to another type of epigenetic alteration. MGMT protein expression may also be reduced due to increased levels of a microRNA that inhibits the ability of the MGMT mRNA to produce the MGMT protein. Zhang (et al.) found, in the glioblastomas without methylated MGMT promoters, that the level of microRNA miR-181d is inversely correlated with protein expression of MGMT and that the direct target of miR-181d is the MGMT mRNA 3'UTR (the three prime untranslated region of MGMT messenger RNA).Epigenetic reductions in expression of another DNA repair protein, ERCC1, were found in an assortment of 32 gliomas. For 17 of the 32 (53%) of the gliomas tested, ERCC1 protein expression was reduced or absent. In the case of 12 gliomas (37.5%) this reduction was due to methylation of the ERCC1 promoter. For the other 5 gliomas with reduced ERCC1 protein expression, the reduction could have been due to epigenetic alterations in microRNAs that affect ERCC1 expression.When expression of DNA repair genes is reduced, DNA damages accumulate in cells at a higher than normal level, and such excess damages cause increased frequencies of mutation. Mutations in gliomas frequently occur in either isocitrate dehydrogenase (IDH) 1 or 2 genes. One of these mutations (mostly in IDH1) occurs in about 80% of low grade gliomas and secondary high-grade gliomas. Wang (et al.) pointed out that IDH1 and IDH2 mutant cells produce an excess metabolic intermediate, 2-hydroxyglutarate, which binds to catalytic sites in key enzymes that are important in altering histone and DNA promoter methylation. Thus, mutations in IDH1 and IDH2 generate a "DNA CpG island methylator phenotype or CIMP" that causes promoter hypermethylation and concomitant silencing of tumor suppressor genes such as DNA repair genes MGMT and ERCC1. On the other hand, Cohen (et al.) and Molenaar (et al.) pointed out that mutations in IDH1 or IDH2 can cause increased oxidative stress. Increased oxidative damage to DNA could be mutagenic. This is supported by an increased number of DNA double-strand breaks in IDH1-mutated glioma cells. Thus, IDH1 or IDH2 mutations act as driver mutations in glioma carcinogenesis, though it is not clear by which role they are primarily acting. A study, involving 51 patients with brain gliomas who had two or more biopsies over time, showed that mutation in the IDH1 gene occurred prior to the occurrence of a p53 mutation or a 1p/19q loss of heterozygosity, indicating that an IDH1 mutation is an early driver mutation.Pathophysiology
High-grade gliomas are highly vascular tumors/tumours and have a tendency to infiltrate diffusely. They have extensive areas of necrosis and hypoxia. Often, tumor growth causes a breakdown of the blood–brain barrier in the vicinity of the tumor. As a rule, high-grade gliomas almost always grow back even after complete surgical excision, so are commonly called recurrent cancer of the brain.Conversely, low-grade gliomas grow slowly, often over many years, and can be followed without treatment unless they grow and cause symptoms.
Several acquired (not inherited) genetic mutations have been found in gliomas. Tumor suppressor protein 53 (p53) is mutated early in the disease. p53 is the "guardian of the genome", which, during DNA and cell duplication, makes sure the DNA is copied correctly and destroys the cell (apoptosis) if the DNA is mutated and cannot be fixed. When p53 itself is mutated, other mutations can survive. Phosphatase and tensin homolog (PTEN), another tumor suppressor gene, is itself lost or mutated. Epidermal growth factor receptor (EGFR), a growth factor that normally stimulates cells to divide, is amplified and stimulates cells to divide too much (EGFR是一种在正常生理下刺激细胞分裂的生长因子,但被放大后,会刺激细胞过多地分裂). Together, these mutations lead to cells dividing uncontrollably, a hallmark of cancer. In 2009, mutations in IDH1 and IDH2 were found to be part of the mechanism and associated with a less favorable prognosis.
IDH1 and IDH2-mutated gliomaPatients with glioma carrying mutations in either IDH1 or IDH2 have a relatively favorable survival, compared with patients with glioma with wild-type IDH1/2 genes. In WHO grade III glioma, IDH1/2-mutated glioma have a median prognosis of ~3.5 years, whereas IDH1/2 wild-type glioma perform poor with a median overall survival of c. 1.5 years. In glioblastoma, the difference is larger. There, IDH1/2 wild-type glioblastoma have a median overall survival of 1 year, whereas IDH1/2-mutated glioblastoma have a median overall survival of more than 3 years.
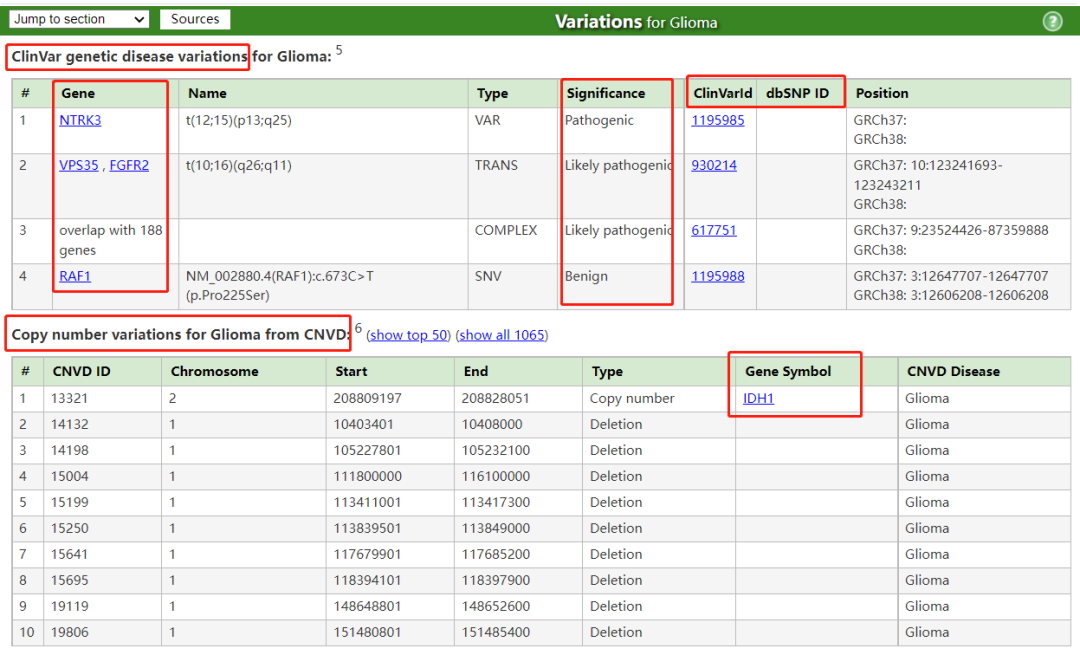
胶质瘤相关基因 (Genes for Glioma, from MalaCards) Genes related to Glioma (11 elite genes): (showing 119, show less) 。星号- Elite gene CC - Cancer Census gene in COSMIC
https://www.malacards.org/card/glioma?limit[RelatedDiseases]=1353&limit[RelatedGenes]=119#RelatedGenes-table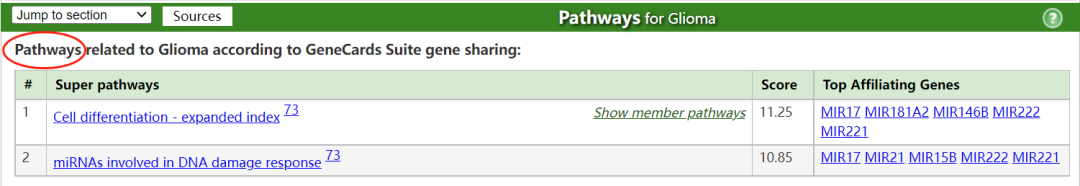
1. 将表格复制、粘贴至Excel表格;2. 另存为“制表符分隔.txt”文件;其它步骤如下:
# 3. 用sed命令删去上肩号Msed -i 's/\r//g' MalaCards-癌种相关的变异.txtsed -i 's/\r//g' MalaCards-癌种相关的通路.txtsed -i 's/\r//g' MalaCards-癌种相关的基因.txt# 4. 确认是否仍存在肩号M,其它特殊符号一般无影响cat -A MalaCards-癌种相关的变异.txtcat -A MalaCards-癌种相关的通路.txtcat -A MalaCards-癌种相关的基因.txt# 5. 获得Gene Listcut -f 2 MalaCards-癌种相关的基因.txt | sort -ur \ > MalaCards-癌种相关的基因.List.txtcut -f 5 MalaCards-癌种相关的通路.txt | \ sed 's/?/\n/g' | sort -ur > MalaCards-癌种相关的通路.List.txtcut -f 7 MalaCards-癌种相关的变异.txt | \ sort -ur > MalaCards-癌种相关的变异.List.txt# 6. 合并Gene Listcat MalaCards-癌种相关的基因.List.txt MalaCards-癌种相关的通路.List.txt MalaCards-癌种相关的变异.List.txt \ > MalaCards-癌种相关的基因-通路和变异.List.txt
链接:https://pan.baidu.com/s/1K16qbB3DGZt3xRJns0Dj2Q 根据上述全文收集到的脑胶质瘤中已被报道的相关基因,为Maf文件取子集 (awk命令):
awk 'BEGIN{OFS=FS="\t"}ARGIND==1{gen[$1]=1}ARGIND==2{if(gen[$1]!="" || FNR==1) print $0}' \ MalaCards-癌种相关的基因-通路和变异.List-Plus-TERT-BRAF-H3K27M-MIR181D-ERCC1-ERCC2-XRCC1.txt \ all.Tumor_Sample_BarcodeAsSampleID.maf \ > all.Tumor_Sample_BarcodeAsSampleID.MalaCards-Glioma-Gene-Term-Variation-Plus-TERT-BRAF-H3K27M-MIR181D-ERCC1-ERCC2-XRCC1.maf
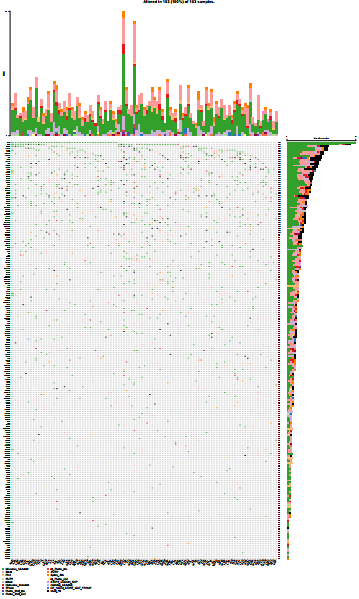
上面的awk命令生成的.maf文件,可使用maftools R包绘制突变瀑布图,网上很多相关R代码,这里不再描述。生成的Maf图概况如下: 结果看起来还不错 (TP53和EGFR都很靠前),这证明了: 当前整个肿瘤WES的分析流程走到这里时,基本的框架和方法是没有问题的。 此外发现一个有趣的结果:TP53的突变所涉及的样本数不是最多的,有一个基因稍稍领先于它,暗示着TP53并不是在所有类型的肿瘤中都是最重要的那一个,而这无疑可能具有重要意义。 某类型的肿瘤中,相关突变基因的完整集合如何获取?可查看此篇文章,否则你的Maf图可能惨不忍睹。
本篇所描述的方法未带有明显的偏见和主观性,而是一种“Knowledge based” (即基于知识库)的筛选模式。尽管此种方法略显繁琐,但是仍可以在任何其它类型的肿瘤中复现,以自行收集所研究的肿瘤中已被报道的所有的突变基因,你可以认为这就是你的大Panel。往期精品(点击图片直达文字对应教程)
机器学习
后台回复“生信宝典福利第一波”或点击阅读原文获取教程合集